UAE
# 10,506
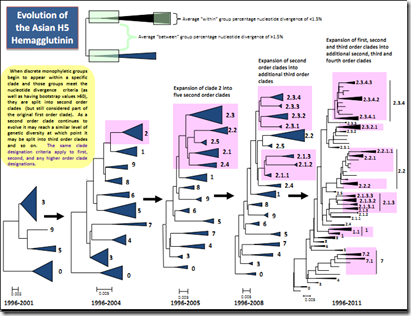
After its initial surge in 2006-2007 – at one point showing up in more than 60 countries – the H5N1 virus began a slow retreat and by 2010 was pretty much relegated to a dozen or so countries where it stayed well entrenched. What it lost in geographic range, it made up for with diversity, as over the years the H5N1 virus has evolved into numerous clades, sub-clades, and variants.
(click to load larger image) (Note: Chart only goes through 2011, and not all clades continue to circulate)
So while we often talk about H5N1 as a single entity, in truth it now encompasses a large family of viruses with varying degrees of infectivity and pathogenicity (see Differences In Virulence Between Closely Related H5N1 Strains). These clades are constantly evolving, and reassorting, and so new versions of H5N1 are continually appearing.
Some are destined to fade away, while others being more biologically `fit’ are able to successfully compete, and thrive.
Over the past two years, the H5N1 virus and its progeny have seen a striking renaissance, with recently emerged clades and their reassorted subtypes (H5N8, H5N2, H5N3, H5N6) spreading with renewed vigor around the globe. Nations that have not seen outbreaks in poultry, or wild birds, since the last decade are now reporting sporadic HPAI H5 once again.
Even more impressively, last fall HPAI H5 has made it to North America for the first time. H5 is clearly on the move again, and helping to drive this revived expansion are two relatively new clades; 2.3.2.1c and 2.3.4.4.
- Clade 2.3.4.4 includes subtypes A(H5N1), A(H5N2), A(H5N6) and A(H5N8), HPAI viruses that have spread rapidly in China over the past couple of years, and some have migrated to Europe and North America.
- Clade 2.3.2.1c has been showing up in Vietnam, China, India, Bulgaria, and Indonesia for several years, and was recently detected in Western Africa This clade was isolated from a nurse who returned to Alberta, Canada from a trip to China (see Alberta Canada Reports Fatal (Imported) H5N1 Infection) in late 2013, and similar to one that killed a captive tiger in Jiangsu Province in 2013.
Just last month we looked at an outbreak of clade 2.3.2.1c last January at Sanmenxia reservoir in Henan province, where scores of migratory birds suddenly died (see Novel H5N1 Reassortment Detected In Migratory Birds – China). Another outbreak occurred in the same region last July, but we have no information on the clade.
Worth noting since in 2005, and again in 2009, large outbreaks of emerging clades of H5N1 in the Qinghai Lakes region of China presaged major expansions of the virus (see 2011 EID Journal New Avian Influenza Virus (H5N1) in Wild Birds, Qinghai, China).
Today we’ve a report of clade 2.3.2.1c showing up in hunting falcons in Dubai (and their captive prey) during the fall of 2014. These detections pre-date the recent arrival of clade 2.3.2.1c in Western Africa, and help to plot the westward expansion of this new HPAI clade.
Authors: Mahmoud M Naguib1, Jörg Kinne2, Honglin Chen3, Kwok-Hung Chan4, Sunitha Joseph5, Po-Chun Wong6, Patrick CY Woo7, Renate Wernery8, Martin Beer9, Ulrich Wernery10, Timm C Harder11
Published Ahead of Print: 07 September, 2015 Journal of General Virology doi: 10.1099/jgv.0.000274
Published Online: 07/09/2015
Highly pathogenic avian influenza viruses (HPAIV) of subtype H5N1 have continued to perpetuate with divergent genetic variants in poultry within Asia since 2003. Further dissemination of Asian origin-derived H5 HPAI viruses to Europe, Africa and, most recently, to the North American continent, have occurred.
We report an outbreak of HPAI H5N1 virus among falcons kept for hunting and other wild bird species bred as falcon prey from Dubai, United Arab Emirates, during fall 2014. The causative agent was identified as avian influenza virus H5N1 subtype, clade 2.3.2.1c, by genetic and phylogenetic analyses. High mortality in infected birds was in accordance with systemic pathomorphological and histological alterations in affected falcons. Genetics analysis showed the HPAI H5N1 of clade 2.3.2.1c is a reassortant in which the PB2 segment was derived from an Asian origin H9N2 virus lineage.
The Dubai H5N1 viruses were closely related to contemporary H5N1 HPAI viruses from Nigeria, Burkina-Faso, Romania and Bulgaria. Median-joining network analysis of 2.3.2.1c viruses revealed that the Dubai outbreak was an episode of a westward spread of these viruses on a larger scale from unidentified Asian sources. The incursion into Dubai, possibly via infected captive hunting falcons returning from hunting trips to central Asian countries, preceded outbreaks in Nigeria and other Western African countries.
The alarmingly enhanced geographic mobility of clade 2.3.2.1.c and clade 2.3.4.4 viruses may represent another wave of transcontinental dissemination of Asian origin HPAIV H5 viruses, such as the outbreak at Qinghai lake caused by the clade 2.2 ("Qinghai" lineage) in 2005.
Thus far H5N1 and its reassortant progeny have failed to adapt well enough to human physiology to pose more than a minor public health threat, although 2015 has already set new records in the number of human infections (mostly in Egypt). Reassuringly, almost all cases have been linked to close contact with infected poultry.
But the rapidly increasing diversity and geographic spread of these HPAI viruses is a concern. One that was voiced quite strongly last spring by the World Health Organization, which warned that H5 is Currently The Most Obvious Avian Flu Threat.
Which makes the tracking, and classification, of avian flu strains more than just of academic interest.










 src="https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEh4zgoKkY5esDyGDfXmhp5tz0W8H2jEgsRJx2wm9317hpr6CTdO8i4DPQj5mF-OAprw6GVcNt84Pt9Yp5U6XEz5h_pAP7azclFEO7kSUzDjr31IvLdzT01usqHnjVk1bBWsqpHQX6G4AIU/s1600/Photo0783.jpg" />
src="https://blogger.googleusercontent.com/img/b/R29vZ2xl/AVvXsEh4zgoKkY5esDyGDfXmhp5tz0W8H2jEgsRJx2wm9317hpr6CTdO8i4DPQj5mF-OAprw6GVcNt84Pt9Yp5U6XEz5h_pAP7azclFEO7kSUzDjr31IvLdzT01usqHnjVk1bBWsqpHQX6G4AIU/s1600/Photo0783.jpg" />












0 komentar:
Posting Komentar